Education:
Ph.D.: 1983, Kansas State University
Pre-OUHSC:
University of North Carolina at Chapel Hill; The State University of New York at Buffalo
Research Interests:
Microbial genomics, bacterial iron transport, bacterial regulatory networks, microbial pathogenesis
Teaching:
Molecular Microbiology, Microbial Pathogenesis, Medical Microbiology, Dental Microbiology
Contact Information:
Office: BRC 1106
Email: David-Dyer@ouhsc.edu
Research Interests:
Microbial genomics
One of our main interests is in microbial genomics. We have participated in several microbial genome sequencing projects, including Neisseria gonorrhoeae, Aggregatibacter actinomycetemcomitans, Actinobacillus pleuropneumoniae, Edwardsiella ictaluri, Flavobacterium columnare, non-typeable Haemophilus influenzae, Histophilus somni (formerly Haemophilus somnus) and Bacillus thuringiensis. We have sequenced the genomes of several Escherichia coli strains responsible for causing neonatal sepsis, in collaboration with Dr. Susana Chavez-Bueno (Children’s Mercy Hospital and University of Missouri at Kansas City). In an ongoing collaboration with Dr. James Chodosh (Massachusetts Eye and Ear and Harvard Medical School), we are sequencing and analyzing the genomes of multiple isolates of Type D human adenoviruses, many of which are associated with epidemic keratoconjunctivitis, an ocular infection.
Regulation of gonococcal gene expression in response to iron availability
A second related interest is microbial pathogenesis. Our laboratory has for some time been interested in the ability of pathogenic bacteria to obtain iron during growth in the human host. Current studies focus on N. gonorrhoeae. The gonococcus can colonize almost any mucosal surface in the human body, although the organism typically infects urogenital mucosae. Our current studies focus on the role of iron as an environmental signal controlling global gene expression responses. Microarray analyses have demonstrated that iron availability directly or indirectly controls expression of ~20% of the gonococcal genome. Direct control of gene expression in response to iron is in part due to the action of the Fur (ferric uptake regulation) protein, as in many other bacteria. However, our data suggest that at least 11 other regulatory proteins and at least one sRNA (nrrF) also participate in controlling the gonococcal iron response. Whole transcriptome and ChIP-seq analyses have allowed us to identify the overall structure of the gonococcal iron response regulon, and the contribution of the Fur regulator to this regulon. We have recently concluded a survey of iron-regulated regulatory sRNAs produced by the gonococcus, and have identified a suite of six sRNAs that appear to be hyper-expressed as the organism enters stationary phase (see Figure 1). The abundance of several of these sRNAs appears to be dependent on the NrrF sRNA, suggesting that these sRNAs represent a regulatory network that may be involved in controlling aspects of gonococcal biology during the unique metabolism of stationary phase. Examining the role of other regulatory factors in the gonococcal iron response continues.
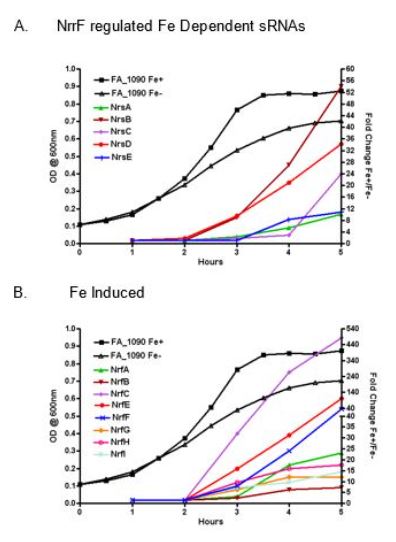
Figure 1: Temporal control of Fe regulated sRNAs. Real time PCR fold change results for all Fe-induced sRNAs were plotted against a representative example of an FA 1090 growth curve showing O.D. values across the 5hr time point to demonstrate the temporal increases in Nrs sRNA A) and Nrf sRNAs B). NrfD is not presented here as this sRNA was Fe-repressed and did not demonstrate temporal control. (Jackson et al., BMC Genomics 18: 317, 2017)
Current Lab Personnel:
Michael Day, Research Assistant III
Lydgia Jackson, Ph.D., Research Assistant Professor
Edgar Scott, Bioinformatics Analyst
Selected Publications:
- Chavez-Bueno S, M. Day M, I. Toby, D. Akins, and D.W. Dyer. May/June 2014. Genome Sequence of SCB34, a Sequence Type 131, Multidrug-Resistance Escherichia coli Isolate Causing Neonatal Early-Onset Sepsis. Genome Announc. 2(3):00514-14. doi:10.1128/genomeA.00514-14. PMID: 24926049
- Day M, Ibrahim M, Dyer D, Bulla L, Jr. July/August 2014. Genome sequence of Bacillus thuringiensis subsp. kurstaki strain HD-1. Genome Announc. 2(4):e00613-14. doi:10.1128/genomeA.00613-14. PMID: 25035322
- Toby, I., and D.W. Dyer. Divergence of protein-coding capacity and regulation in the Bacillus cereus sensu lato group. BMC Bioinformatics 2014, 15(Suppl 11):S8 (21 October 2014) PMID: 25350501
- Day, M.W., L.A. Jackson, D.R. Akins, D.W. Dyer and S. Chavez-Bueno. Whole Genome Sequences of the Archetypal K1 Escherichia coli Neonatal Isolate RS218, and the Contemporary Neonatal Bacteremia Isolates SCB11, SCB12, and SCB15. Genome Announc. January/February 2015 3:e01598-14; doi:10.1128/genomeA.01598-14. PMID: 25720688
- Ramke, M., J.Y. Lee, D.W. Dyer, D. Seto, J. Rajaiya, and J. Chodosh. The 5’ UTR of human adenoviruses: new insights into evolution and leader diversity. Virology. 2015 Nov;485:452-9. doi: 10.1016/j.virol.2015.08.018.
- Singh, G., X. Zhou, J.Y. Lee, M. Yousuf, M. Ramke, M. Ismail, J. Lee, C. Robinson, D. Seto, D. Dyer, M. Jones, J. Rajaiya, and J. Chodosh. Recombination of the Epsilon Determinant and Corneal Tropism: Human Adenovirus Species D types 15, 29, 56, and 69. Virology. 2015 Nov;485:452-9. doi: 10.1016/j.virol.2015.08.018. Epub 2015 Sep 7. PMID: 26343864
- Kenedy, M.R., E.J. Scott II, B. Shrestha, A. Anand, H. Iqbal, J.D. Radolf, D.W. Dyer and D.R. Akins. Consensus Computational Network Analysis for Identifying Candidate Outer Membrane Proteins from Borrelia Spirochetes. BMC Microbiology BMC Microbiology 16: 141 (2016); DOI 10.1186/s12866-016-0762-z.
- Ismail, A.M., J. Lee, D.W. Dyer, D. Seto, J. Rajaiya and J. Chodosh. Selection Pressure in the Human Adenovirus Fiber Knob Drives Cell Specificity in Epidemic Keratoconjunctivitis. J Virol. 2016 Oct 14;90(21):9598-9607. Print 2016 Nov 1.
- Chavez-Bueno, S., B. coli, M. Iliki, D. Akins, D. Dyer, D. Bard and E. Scott II. Route of Infection Alters Virulence of Neonatal Septicemia Escherichia coli Clinical Isolates. PLOS ONE, December 13, 2017 https://doi.org/10.1371/journal.pone.0189032
- Jackson, L., M. Day, J. Allen, E. Scott II, and D.W. Dyer. Iron-Regulated Small RNA Expression as Neisseria gonorrhoeae FA_1090 Transitions into Stationary Phase Growth. BMC Genomics 18: 317, 2017; DOI: 10.1186/s12864-017-3684-8.
- Day, M., E. Scott II, L. Jackson and D.W. Dyer. ChIP-seq analysis of Fur binding and the Iron Response Regulon of Neisseria gonorrhoeae FA1090. BMC Genomics (submitted).
- Jackson, L.A., M. Day, E. Scott II, J. Allen and D.W. Dyer. Transcriptome analysis of Neisseria gonorrhoeae FA1090 grown under iron-limiting conditions. BMC Genomics (submitted).